Target Identification with informatics and data mining
What is druggability?
Central to the drug discovery process is the identification of a suitable efficacious target, and the ability of a novel drug-like compound to bind to and modify that target. A successful drug target needs to demonstrate two key properties:
- it has to have a site capable of binding drug-like molecules, i.e. a druggable site, and
- it has to have a causal link to a disease process. Historical drug targets have both of these properties by definition, and analysis of their features can help guide and derisk future drug discovery.
Druggability usually runs in families
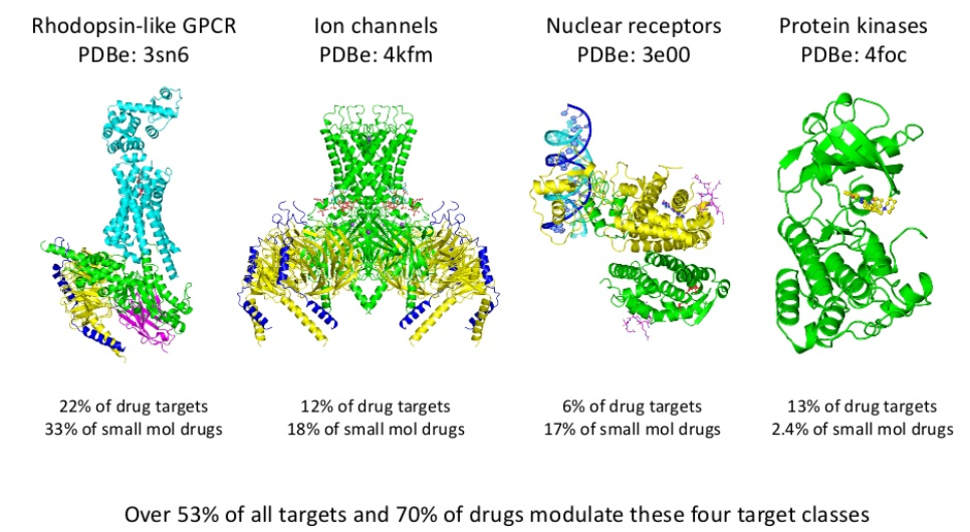
The curation and analysis of databases of known drug targets have allowed them to be classified into protein families, within which are four main target classes of privileged ‘druggable’ families – Rhodopsin-like GPCR ligands, ion channels, nuclear receptors and protein kinases. Approximately 53% of historical drug targets and 70% of approved drugs modulate one of these 4 targets, so investing in screening technologies, in compound libraries and in expertise around the system biology and signalling of these proteins supports the drug discovery process, alongside the use of informatics to gather data on the desired target.
Use of these databases, i.e. the ChEMBL database, shows known drug targets such as GPCR ligands, yield a good return on investment – 18% of compounds published in lead optimisation studies are GPCR ligands, while 30% of approved drugs on the market are GPCR drugs. So whilst identification of a novel drug target is scientifically fascinating and exciting, discovery productivity will be lower with a significant investment in time and resource required as the explicit cost of this higher novelty.
Foresight via genetics
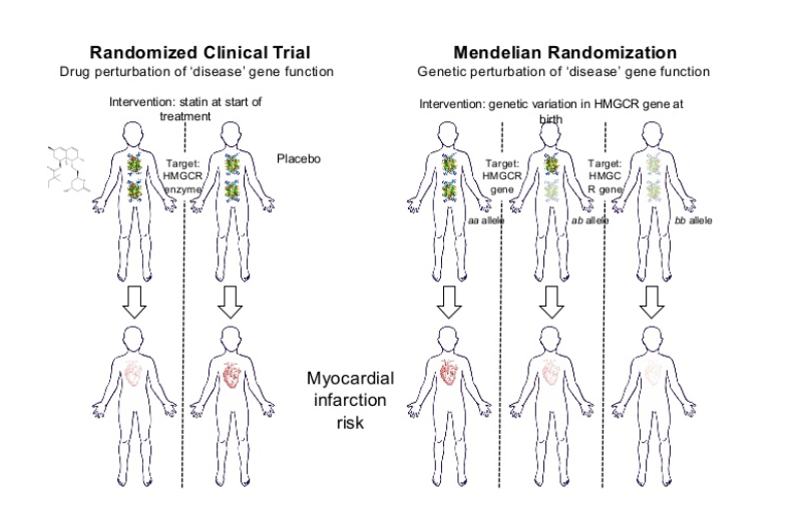
Whilst knowledge of target druggability is essential, so is the efficacy component. Mendelian randomisation can provide evidence of the causal relationship between the target and disease and provide a way of anticipating the likely success of a target. The real benefit of pre-validating the success of these targets is it can be done prior to large scale, expensive phase 2 trials.
Using the resources available
When considering drug targets for analysis, triage and so forth, the use of online resources can support these decisions. Examples include Open Target – a collaborative project between several industry partners, the EMBL-EBI and the Sanger Institute who publish a richly curated and integrated collection of data. Illuminating the Druggable Genome (IDG) is a global project whose aim is to identify and provide information on less well studied proteins within commonly drug-targeted protein families. Finally, the CanSAR platform at the Institute of Cancer research provide data on somatic diseases.
This article is based on Davide’s talk from the MDC Connects webinar series. Watch the session John took part in – Identifying the Target:

About the author
Professor John Overington joined Medicines Discovery Catapult as CIO in 2017, where he leads the development and application of informatics approaches to promote and support innovative, fast-to-patient drug discovery in the UK through collaborative projects across the applied R&D community.
John was involved in the development of the medicinal chemistry database StARLite – the precursor to ChEMBL. More recently, the work extended into large-scale patent informatics with the Open patent database SureChEMBL. John has a degree in Chemistry from the University of Bath and a PhD from Birkbeck College, London. He is a visiting professor at UCL and the University of Manchester.