Over the past two decades Matrix Assisted Laser Desorption Ionisation Mass Spectrometry Imaging (MALDI-MSI) has been widely applied in biomedical science and life sciences. MALDI-MSI of biological tissue was initially developed by Richard Caprioli in 1997 following on from the pioneering work of Bernard Spengler. The technology allows for untargeted measurement of peptides and proteins from multiple samples.
MSI uses the sensitivity and selectivity of MS to show the distribution of specific species within tissue sections thus providing chemical composition and spatial information simultaneously. As MSI techniques do not require any specific antibodies or probes, they can provide a wealth of additional knowledge to the field of molecular histology.
FFPE analysis: the common workflow
MSI can be applied to formalin-fixed and paraffin-embedded (FFPE) tissues as well as fresh frozen sections; an important aspect as most tissues with clinical relevance are collected and stored by pathologists as FFPE samples. Held by hospitals and biobanks, FFPE blocks can be stored for many years.
A combination of deparaffination, heat induced antigen retrieval and in situ proteolytic digestion enables the analysis of proteins by MSI; an application first reported by the group of Isabelle Fournier in 2007 (Lemaire et al., 2007) and later defined as ‘the new frontier or histopathology proteomics’ (Fournier et al., 2008).
Following the removal of paraffin, antigen retrieval is carried out to break the methylene crosslinks of proteins, that are formed during the fixation stage, by incubating the glass slide in a buffer at high temperatures. Digesting the now released larger proteins into smaller peptides also helps to counteract the inefficiencies of Time-of-Flight (TOF) mass spectrometers in resolving higher molecular weight compounds. The peptide fragments are much easier to detect and visualize by MALDI-MSI than intact proteins, however the parent protein can still be inferred from the localization of the fragment.
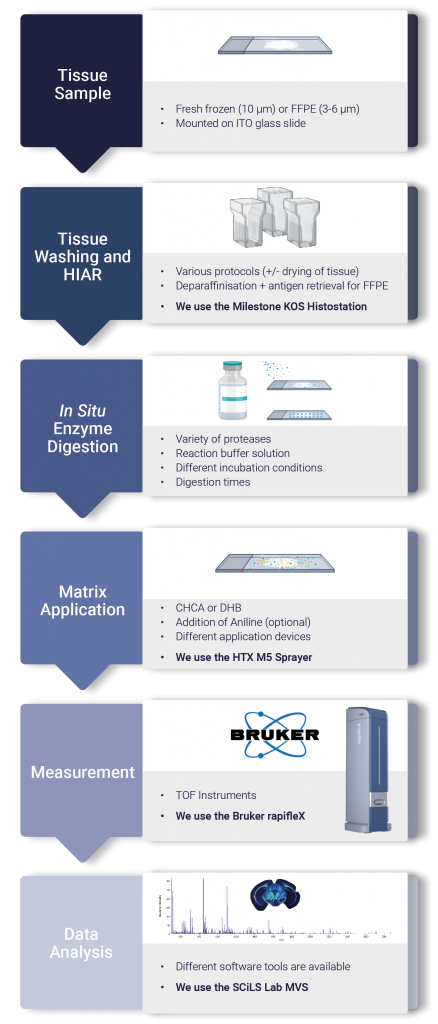
Figure 1. Outlines the key steps in the FFPE analysis workflow. Tissues are sectioned onto ITO coated slides with a microtome at 3-6 µm. Following a series of sequentially decreasing ethanol concentrations the tissue is rehydrated in preparation for HIAR. With the proteins released, a protease such as trypsin can be sprayed (using the HTX M5 sprayer) or spotted onto the glass slide. After incubation in a humid environment the slide is sprayed with matrix and analysed using the Bruker rapifleX.
The importance of developing an FFPE workflow
At Medicines Discovery Catapult, we are working with the latest sample preparation technologies by HTX Imaging (M5 sprayer) to utilise and maximise the full capabilities of the Bruker rapifleX MALDI.
Developing robust and standardised protocols for the analysis of FFPE sections allows us to delve into hundreds and thousands of archived blocks within tissue banks. Archived FFPE samples provide the possibility of retrospective analyses to correlate molecular patterns to patient data thus having the potential to detect markers for disease and treatment responses.
About the author
Dr Ekta Patel, PhD, is a Senior Scientist in Mass Spectrometry at Medicines Discovery Catapult with over 7 years’ experience in the field of imaging, both in academic and industrial roles.
During her PhD, Ekta further developed methods to detect peptide / protein biomarkers from fingermarks and tissue via shotgun in situ proteomics. Other focus projects included working with GlaxoSmithKline as an Industrial PhD Scientist to develop non-invasive methods to monitor patient compliance. Prior to MDC she was a Global Applications specialist for MALDI Mass Spectrometry Imaging at a major Mass Spectrometry vendor, Kratos Analytical Shimadzu.