Aurelia Bioscience are a pre-clinical contract research organisation specialising in the development of bespoke assays and discovery-based projects for clients. The company offers assay development to detect protein-protein interactions in living cells using technology such as the NanoBRET target engagement assay, cell culture using 3D cell-based screening applications and magnetic electrospun micro-fibre technology, the study of protein degradation using proteolysis targeting chimeras (PROTAC) and protein detection and characterisation using Western Assay System (WES/JESS).
Protein-protein interactions in living cells
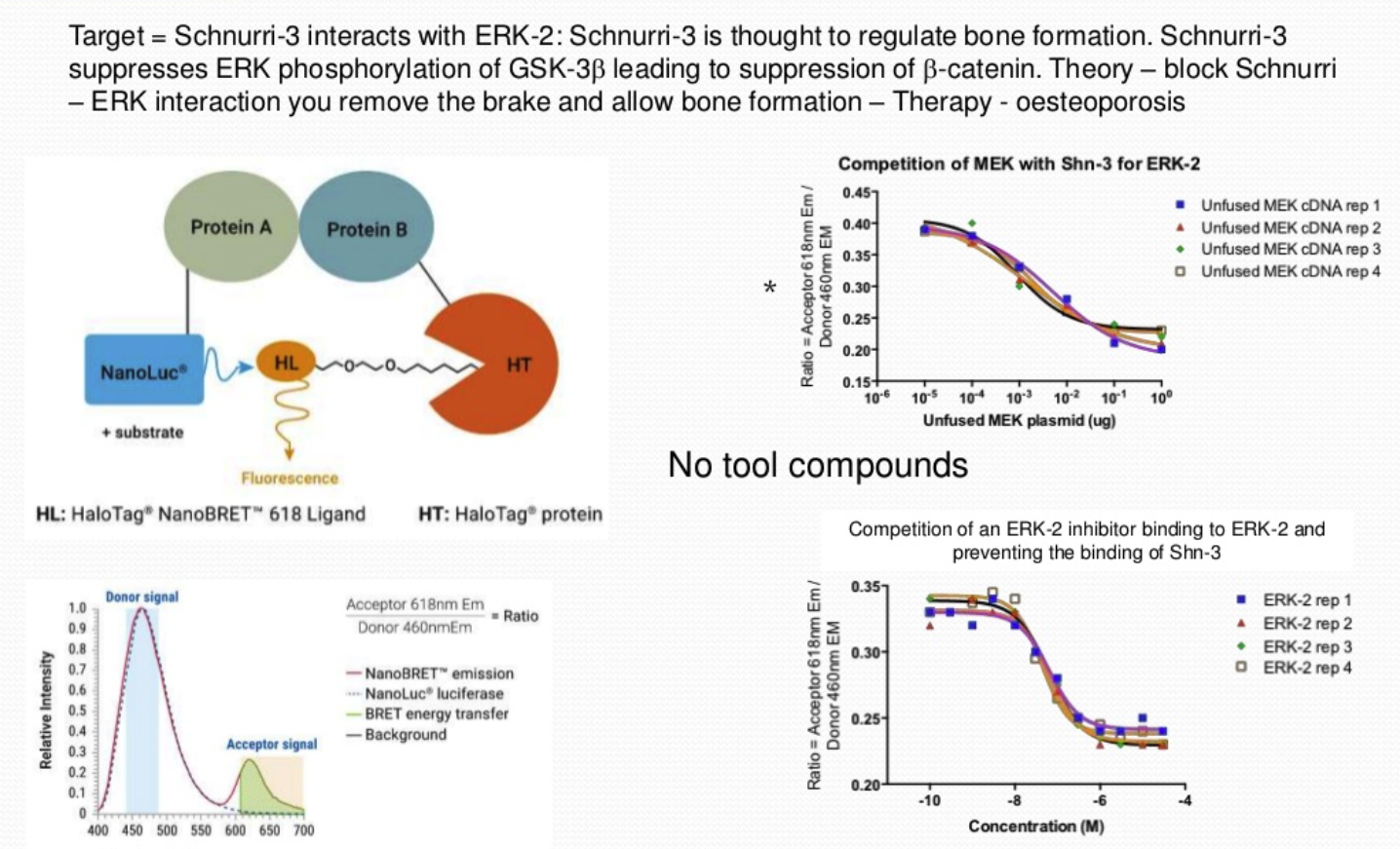
NanoBRET technology studies protein-protein interactions in living cells, using Bioluminescence Resonance Energy Transfer (BRET). A study investigating the interaction of Schnurri-3, a regulator of adult bone formation, with ERK-2 used 2 different vectors. One vector contained Schnurri-3 and NanoLuc®, a luminescence enzyme with a high efficiency to produce photons, and the 2nd vector contained ERK-2 plus HaloTag®, a fluorescent ligand. Fluorescence was observed when the two proteins came into close proximity. The NanoBRET assay screened for inhibitors of the Schnurri-3-ERK interaction and the ratio of the NanoLuc signal to fluorescence signal was determined. This was validated using MEK within the cells which competes for interaction and if over-expressed will decrease the fluorescence signal.
Based on these results, a high throughput screening assay was developed and 50,000 compounds were screened which identified a smaller number of compounds that interacted with Schnurri-3, blocking its binding to ERK-2.
NanoBRET Target Engagement
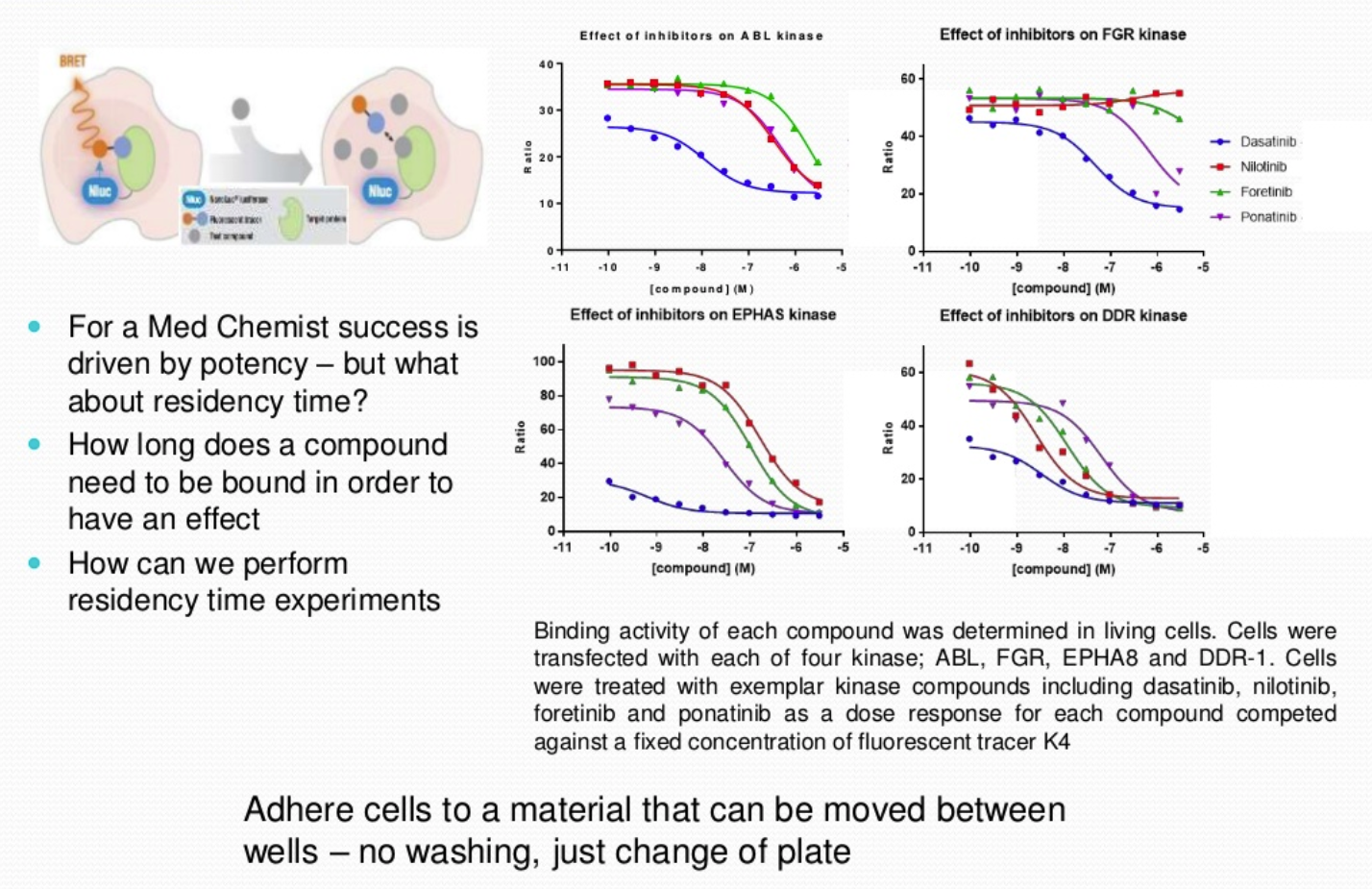
NanoBRET Target Engagement is a competition assay between a fluorescently labelled tracer and a kinase of interest, the interaction occurs in living cells. The full-length kinase is expressed in the cell and interaction takes place at a physiological ATP and physiological pH enabling the association and dissociation rates of the compound within the cells to be observed. The use of a labelled tracer activates fluorescence when in close proximity to the NanoLuc enzyme expressed either N- or C-terminal of the kinase protein, to produce a signal. This signal will decrease if a competing compound is introduced, and the tracer is no longer in close proximity to the NanoLuc enzyme.
The assay also demonstrates both cellular permeability and potency of the compounds.
3-D cell-based screening
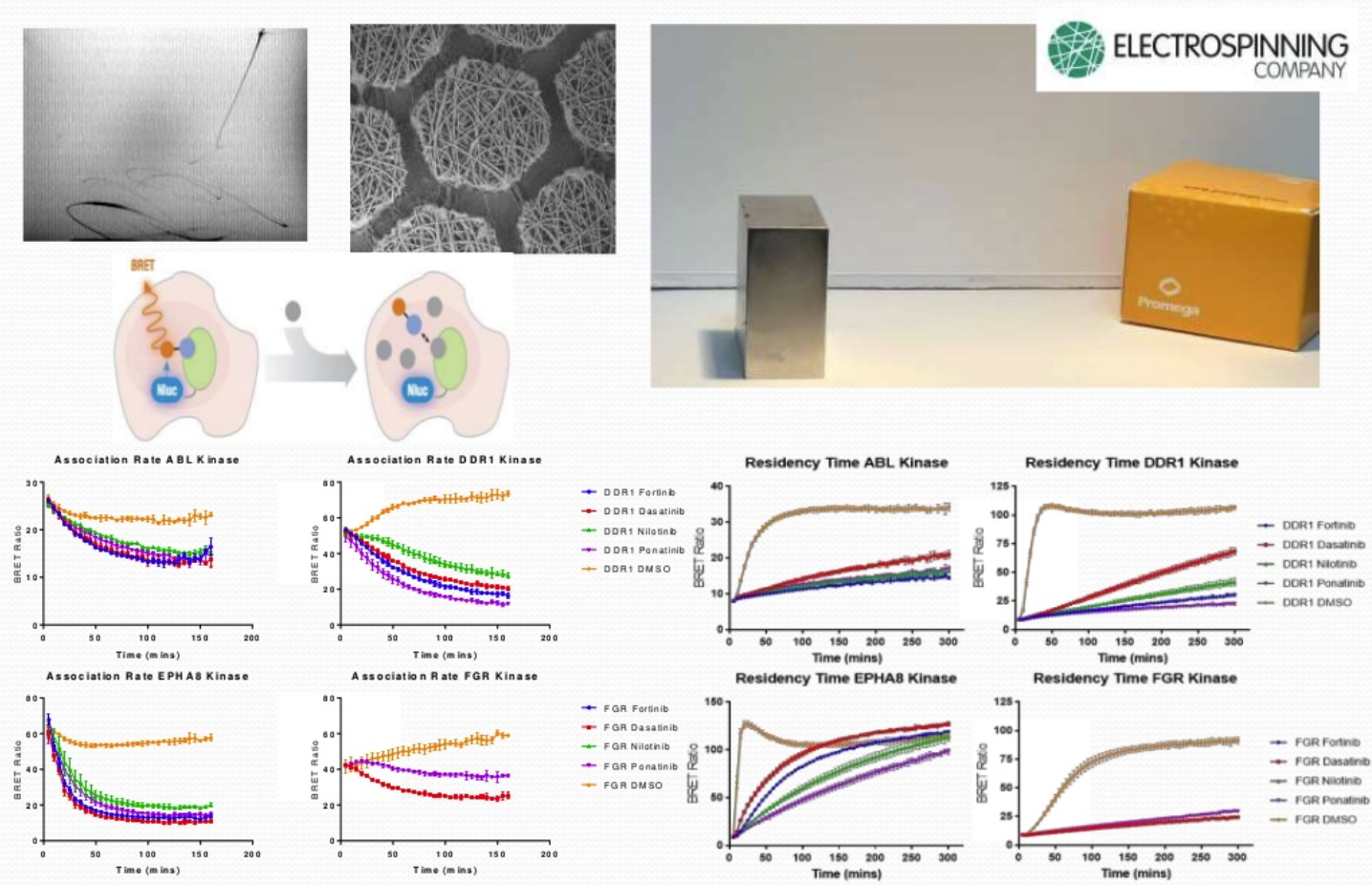
To investigate the residency time of how long a compound needs to be bound to a target to have an effect, an electrospun scaffold material has been developed onto which cells are seeded. The scaffold is magnetic allowed movement of an adherent monolayer of cells from well to well i.e. wells containing the tracer or wells containing compound. These can then be placed directly into the plate reader. The resulting graph shows time vs. the BRET ratio and the different compounds compete off the receptor at different rates.
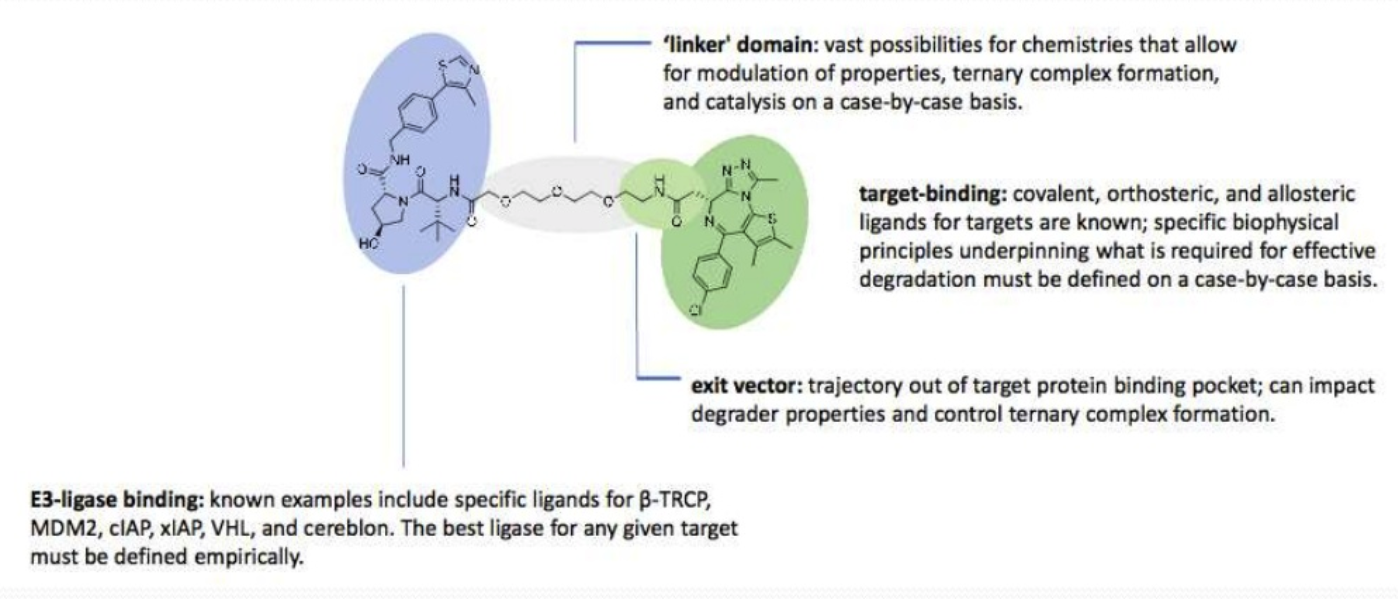
Proteolysis targeted chimeras (PROTAC)
PROTAC uses a ubiquitin-proteasome system to induce protein degradation at the proteasome. The technology uses a small chimeric molecule, one end selectively binds to the protein target of interest and the other end binds to an E3 ligase, held together with a linker. The E3 ligase ubiquitinates the protein, targeting it for degradation. Once degradation is in progress the PROTAC molecules dissociates and can be reused. An advantage of this system is it degrades the target protein rather than inhibits it, making it an excellent technique to study undruggable proteins.
Following treatment with PROTAC, high-throughput, capillary based, automated Western blot, WES and JESS are used to measure protein degradation, Both techniques are fully automated, the cell ligase/protein of interest is separated based on molecular weight, immobilised with UV and subjected to immunoprobe by aspirating the primary antibody, to generate a luminescence read out.
This article is based on Gary’s talk from the MDC Connects webinar series. Watch the session Gary took part in – Hit Identification:

About the author
Gary Allenby is CEO and founder at Aurelia Bioscience where he leverages over 30 years of pre-clinical drug discovery experience and a number of scientific papers in leading journals to aid the discovery of new chemical entities.
After gaining his PhD from Edinburgh University in 1990, Gary joined Hoffmann La Roche (USA) to investigate retinoid pharmacology and the role of nuclear hormone receptors in foetal development. He then went on to work in the Lead Generation Section of GSK (UK) in 1995, gaining expertise in the development of cell-based assays within the CNS disease area for high throughput screening as well as novel drug discovery assay technologies and automation. After leaving GSK, Gary spent over 10 years at AstraZeneca (UK) before founding Aurelia Bioscience in 2011.