Challenges and Opportunities of Complex Cell Models for Toxicity Testing
Availability of safety data is critical in early studies and in later pre-clinical development. A key focus is to improve the safety related attrition in clinical studies which could be achieved using model systems closer to humans and therefore able to generate more reliable, translatable data. These data can then be used to reliably identify toxicity flags throughout discovery.
The current cell models used are often too simple and inadequate to provide these data. Cells are immortalised and have aberrant cellular metabolism, models are often single cell, cultured in a simple 2D setup, used with a glass or plastic substrate, lacking in relevant biological stimuli, with no communication with other cells or cellular models and there is an absence of relevant physical stimuli.
Next generation models accessing stem cells, primary cells with the ability to modify using CRISPR represent more biologically relevant models. Multiple different cell types should be cultured together to mimic in vivo situations, Newer models could be 3D spheroids unsupported structures or with matrix gels, or even patient-derived organoids. Available technologies to help better mimic biological environments should be utilised eg. the presence of flow.
To exploit more complex cell models, it will be necessary to validate and use humanised isolated organ on-chip models (OOAC) and connected on chip models which better mimic a normal biological environment, for use in both efficacy and safety determination.
A key step is to develop isolated models and multiple organ models using new technology and identifying promising models that can be used. Industrial experience can provide the expertise to characterise the model, elucidate how good it is, how robust it is, and identify limitations.
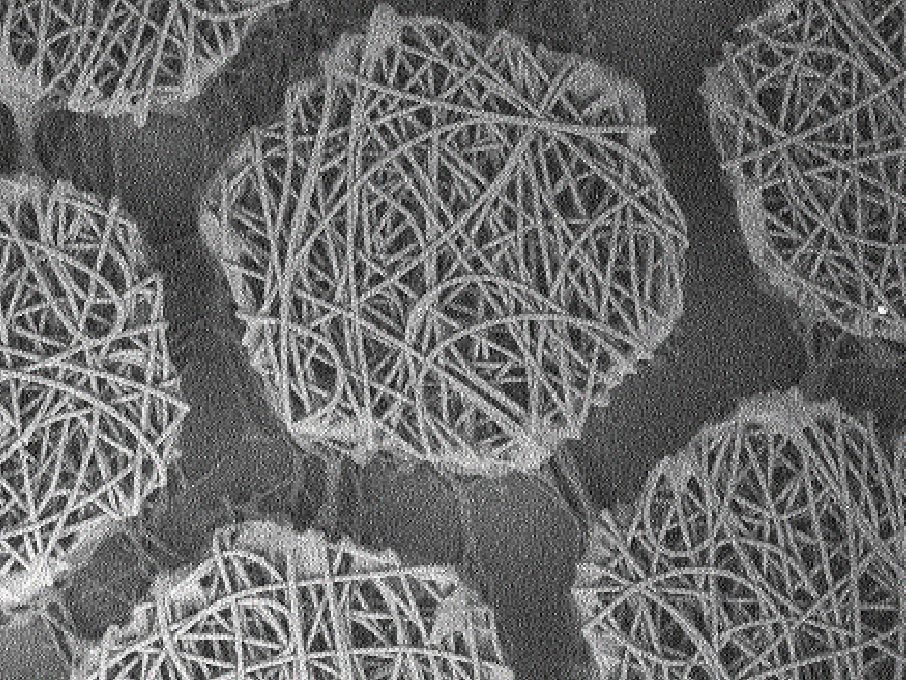
Medicines Discovery Catapult (MDC) are developing and evaluating next generation cell models. Examples include the CMEF Cardiac Model, a 3D model for assessment of cardiotoxicity. It uses stem cell derived cardiomyocytes joined with primary cardiac fibroblasts and cardiac endothelial cells (CMEF). These cells, placed in ultra-low attachment wells of a plate, will form spheroids and will then spontaneously beat. By positioning the cells on a recording electrode of a cardioexcyte instrument, it is possible to determine the beat rate and amplitude and produce a trace.
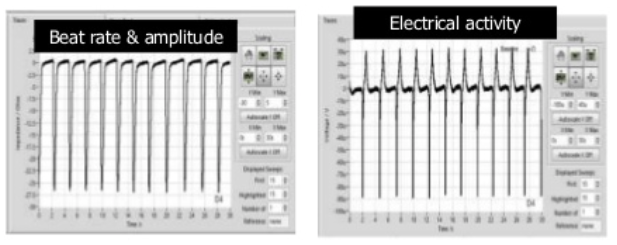
The model has been characterised using known clinically relevant human cardiotoxins and results demonstrated improved predictability, with stepwise increases in predictivie power with the ccuulation of each cell type. The model is amenable to screening at discovery scale, exploits label free, continuous and real-time measurements. There are current ongoing collaborations for CRISPr introduced cardiac mutations and endothelial cell optimisation to further enhance translation to patient physiology.
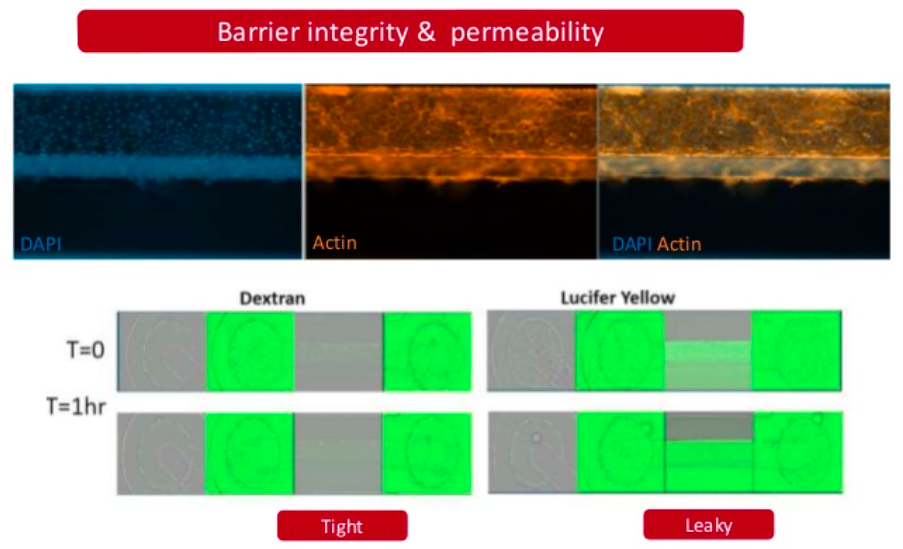
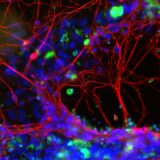
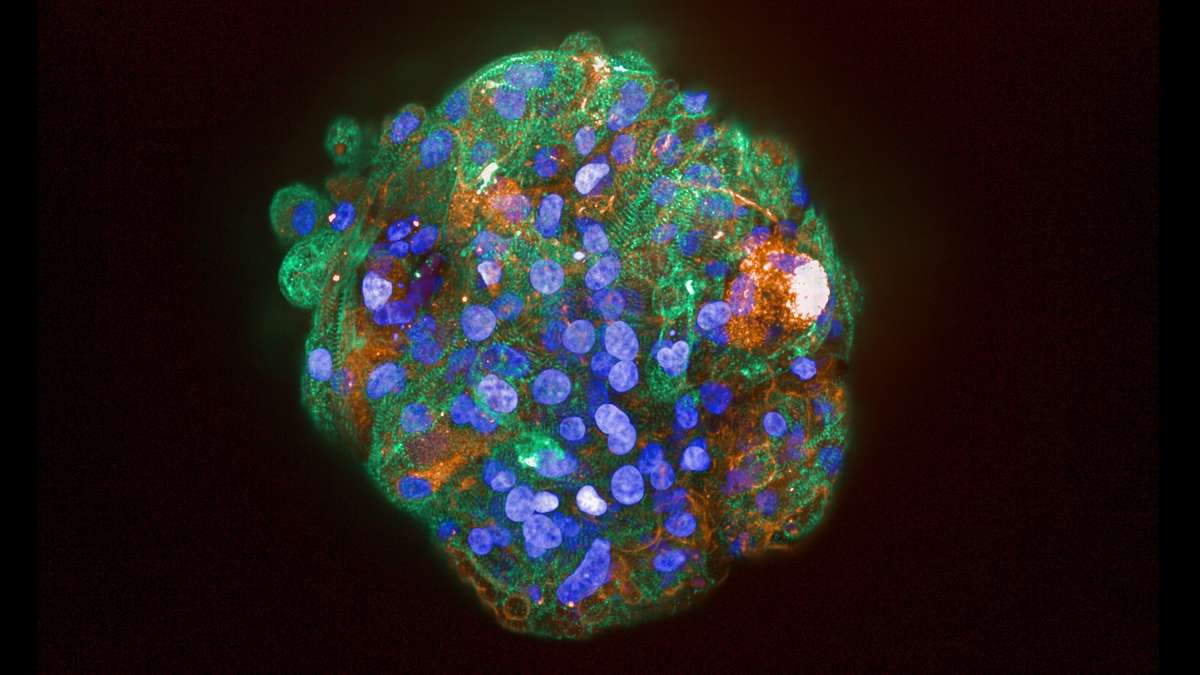
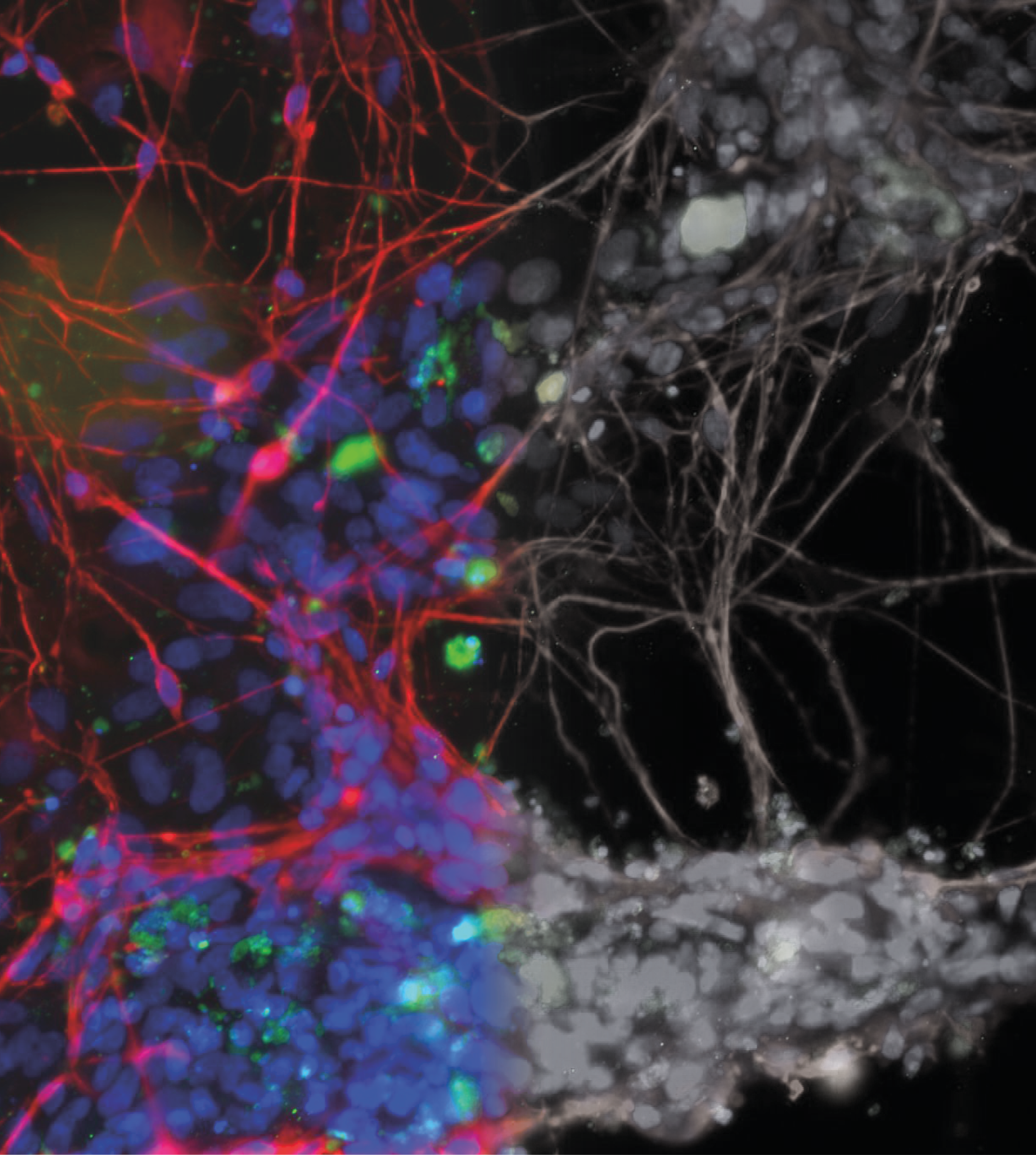
Neurotoxicity is also a significant problem and the blood-brain barrier (BBB) represents an opportunity to produce a model to give more accurate and predictive results. Current standard in vitro BBB models use Caco-2 or MDCK-MDR1 cell lines in transwell plates alongside in vivo rodent models for compound permeability and neurotoxicity, however they have limited physiological relevance. MDC are evaluating a simple BBB model in a Mimetas OOAC -384 well SBS plate format. It has a 96-chip or 40-chip format, and the ability to form a tubule from for example, primary human endothelial brain cells, with a simple flow. It has compartmentalised channels and allows cross-talk between cells. Images below, using a fluorescent dye, show the barrier integrity and permeability – there has been some leakage across the barrier so further optimisation is required of the model.
Highlighted here, are just 2 examples of models being evaluated at MDC, however other models include a liver model using a Physiomimix OOAC platform and primary hepatocytes.
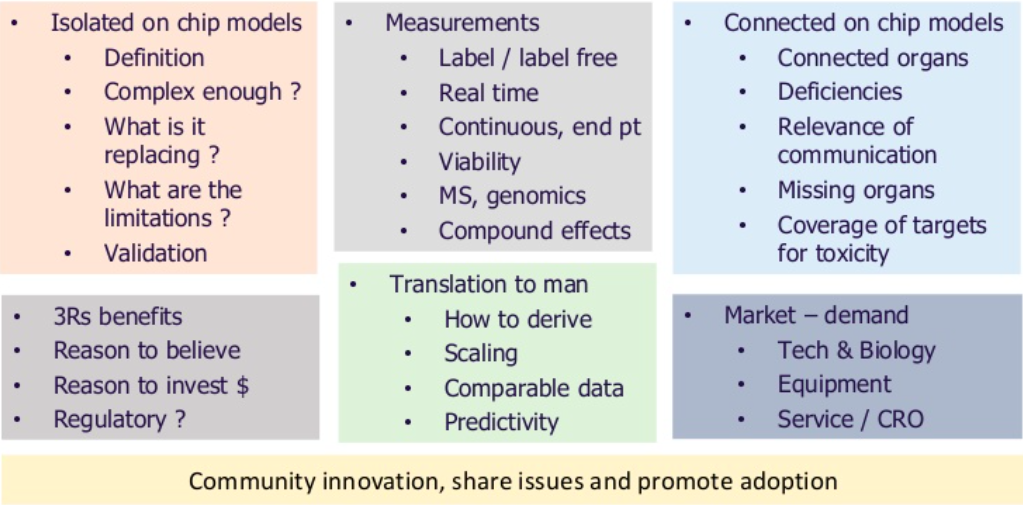
The ultimate aim in next generation model development is to connect together robust and translatable relevant human models. There is currently a lot of activity in the cell nanochip space looking to do this. However, there are major challenges to overcome as shown below.
In order for these next generation models to replace or augment animal studies it is important to understand where they can be positioned in drug discovery for safety testing and the value of the data that they will generate. Progression of these models will involve a collaborative approach across UK academia and drug discovery communities.
This article is based on Malcolm’s talk from the MDC Connects webinar series. Watch the session Malcolm took part in – Is my Compound Safe?:

About the author
Malcolm Haddrick is a Lead Scientist at Medicines Discovery Catapult with specific responsibility for complex cell model development and organ-on-a-chip applications. He has worked across efficacy and toxicity projects at AstraZeneca and Pfizer for the last 20 years, developing and running screening assays for small molecule drug discovery.