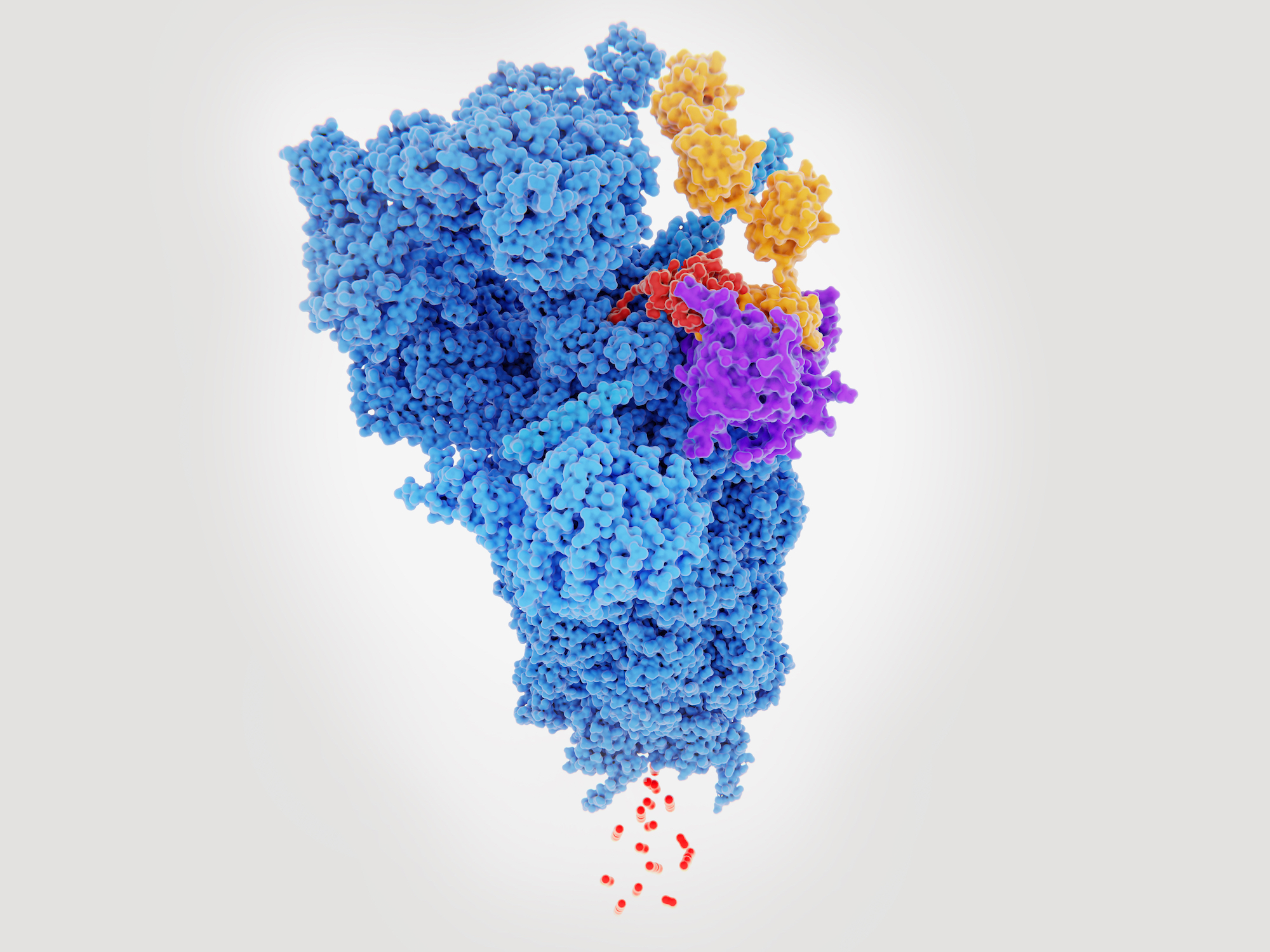
Where can CryoEM be positioned in Medicines Discovery?
Cryo-electron microscopy (cryoEM) is a powerful structural biology technique that can be used to study a range of different macromolecular complexes and questions in medicines discovery.
What is cryo-electron microscopy
Recent advances in both software and hardware in cryoEM mean we can now routinely resolve structures for a whole range of different macromolecular complexes, to resolutions of around 3 angstroms.
CryoEM can be used for structure determination of potential targets, to directly examine the location of a small molecule, or examine binding of antibody or non-antibody binding proteins. Even where there may be heterogeneity (both in terms of compositional and conformational heterogeneity), cryoEM is often able to generate high quality structural information.
Advantages of cryo-EM
Typically, single particle cryoEM can enable structure determination of proteins and macromolecular complexes of ~100 KDa up to 1000s of KDa. CryoEM can provide structural information on large macromolecular complexes with many subunits or complexes with disordered regions, where other structure determining techniques such X-ray crystallography might not be suitable.
CryoEM is also useful for protein complexes where it is difficult to generate sufficient volumes or concentrations for other structural techniques such as X-ray crystallography, as cryoEM requires lower volume and concentrations of material.
How it works
The first stage is to generate a high-quality protein preparation. Negative staining electron microscopy can be used as an initial quality control step to provide an indication of purity, heterogeneity, aggregation, and degradation, to give a good indication of how the sample will behave in single particle cryoEM analysis.
The sample can then be prepared for cryoEM via vitrification. Approximately 3 µL of the macromolecular complex is applied to a cryoEM grid. A thin film (10-100nm) of liquid containing the specimen of interest is then formed and immobilised by plunge freezing into a cryogen such as liquid ethane to rapidly to a vitreous state.
The cryoEM grid is then screened to ascertain if the quality is good enough for a full data collection, This process can be manual, or automated with a small dataset collected. Initial results from a small dataset can be obtained in as little as 30 minutes using new image processing pipelines, which are entirely automated.
During full data collection, the optimised grid is placed into a Titan Krios microscope for data collection, which takes between 24 and 72 hours to generate raw data. We can then use single particle averaging techniques to generate a high-resolution three-dimensional structure of our protein or macromolecule of interest.
CryoEM opens the doors to structure-based drug design approaches to be applied to previously intractable targets. CryoEM is well suited to the analysis of membrane proteins, and other complexes which are hard to work with. Overall, cryoEM is a powerful tool for structure based drug design and medicines discovery.
The University of Leeds can offer expert support and equipment access in cryoEM. We support the full pipeline of structure determination by cryoEM, from sample quality control and preparation, to data collection and full analysis. We also offer comprehensive training opportunities. The facility sits within a wide network of world-class research facilities within the University, enabling us to take on complex and multi-faceted challenges in medicines discovery.
This article is based on Rebecca’s talk from the MDC Connects webinar series. Watch the session Rebecca took part in – Structural Approaches for Drug Discovery:

About the author
Rebecca Thompson is the Electron Microscopy Manager and senior cryo-electron microscopy scientist within the Astbury Biostructure Laboratory, University of Leeds. In this role she oversees the management of the facility, which includes two state-of-the-art Titan Krios microscopes. Her current research interests span cryo-electron microscopy and include developing and optimising workflows for high resolution structure determination of macromolecular complexes by single particle analysis, and using cryo-electron tomography to image cells and organelles.